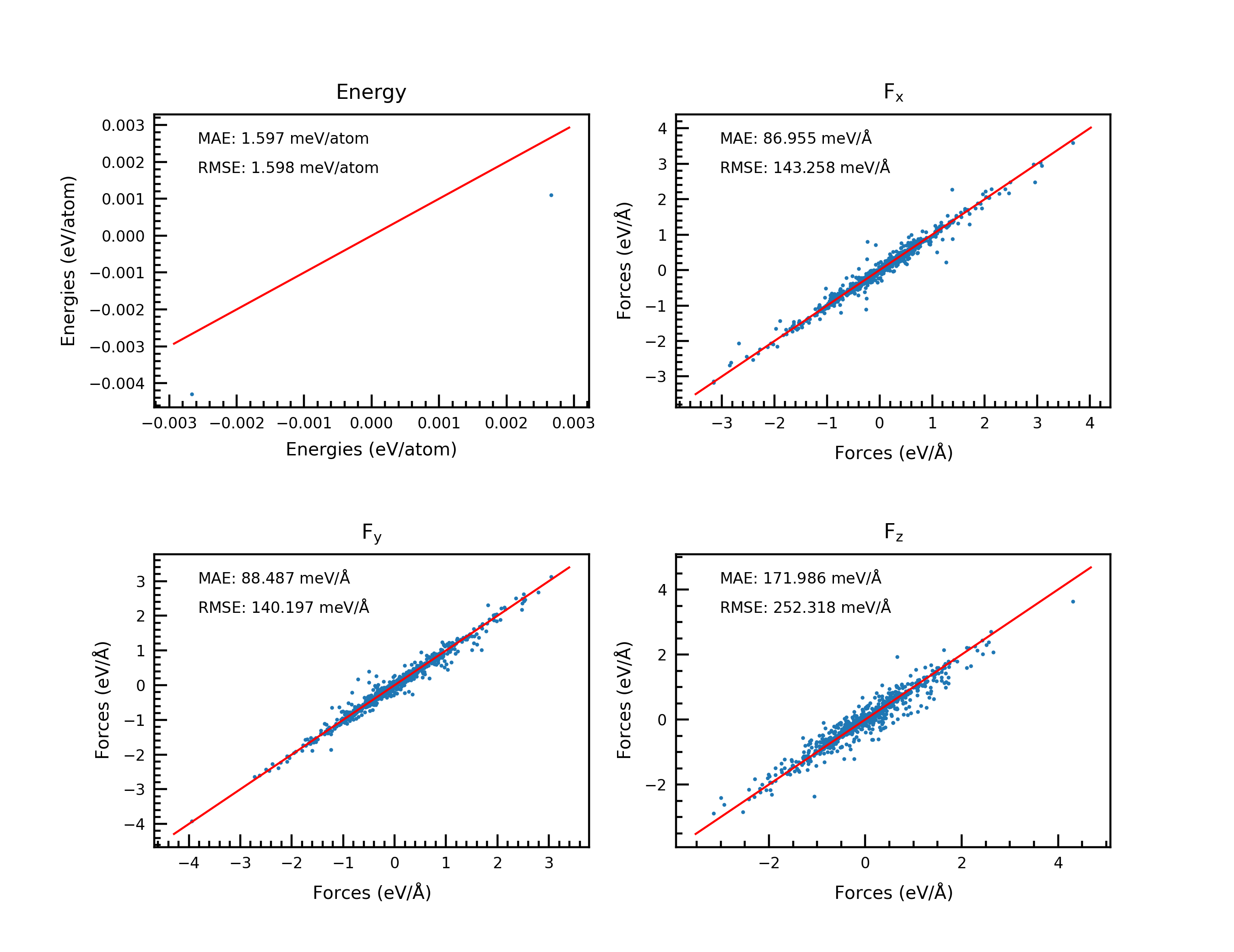
Plot DPMD data#
Plot dptest#
Nowadays, I found that many studies for the simulations of interfaces use DeePMD-kit. In the end, users will need to test the accuracy of the machine learning potentials. Here, I implemented a simple script to plot the results of the tests.
Assume users have had two dptest files from DeePMD-kit, called dptest.e.out and dptest.f.out.
Now we import the plot function from ECToolkits
from ectoolkits.plots.dpmd import plot_dptest
e_file = "./dptest.e.out"
f_file = "./dptest.f.out"
save_png = "dptest.png"
fig = plot_dptest(e_file=e_file, f_file=f_file, save_name=save_png, frc_comp=True, return_err=False)
fig, rmse_e, mae_e, rmse_fx, mae_fx, rmse_fy, mae_fy, rmse_fz, mae_fz = plot_dptest(e_file=e_file, f_file=f_file, save_name=save_png, frc_comp=True, return_err=True)
fig, rmse_e, mae_e, rmse_f, mae_f = plot_dptest(e_file=e_file, f_file=f_file, save_name=save_png, frc_comp=False, return_err=True)
With the above codes, users can get the plot of the test results.
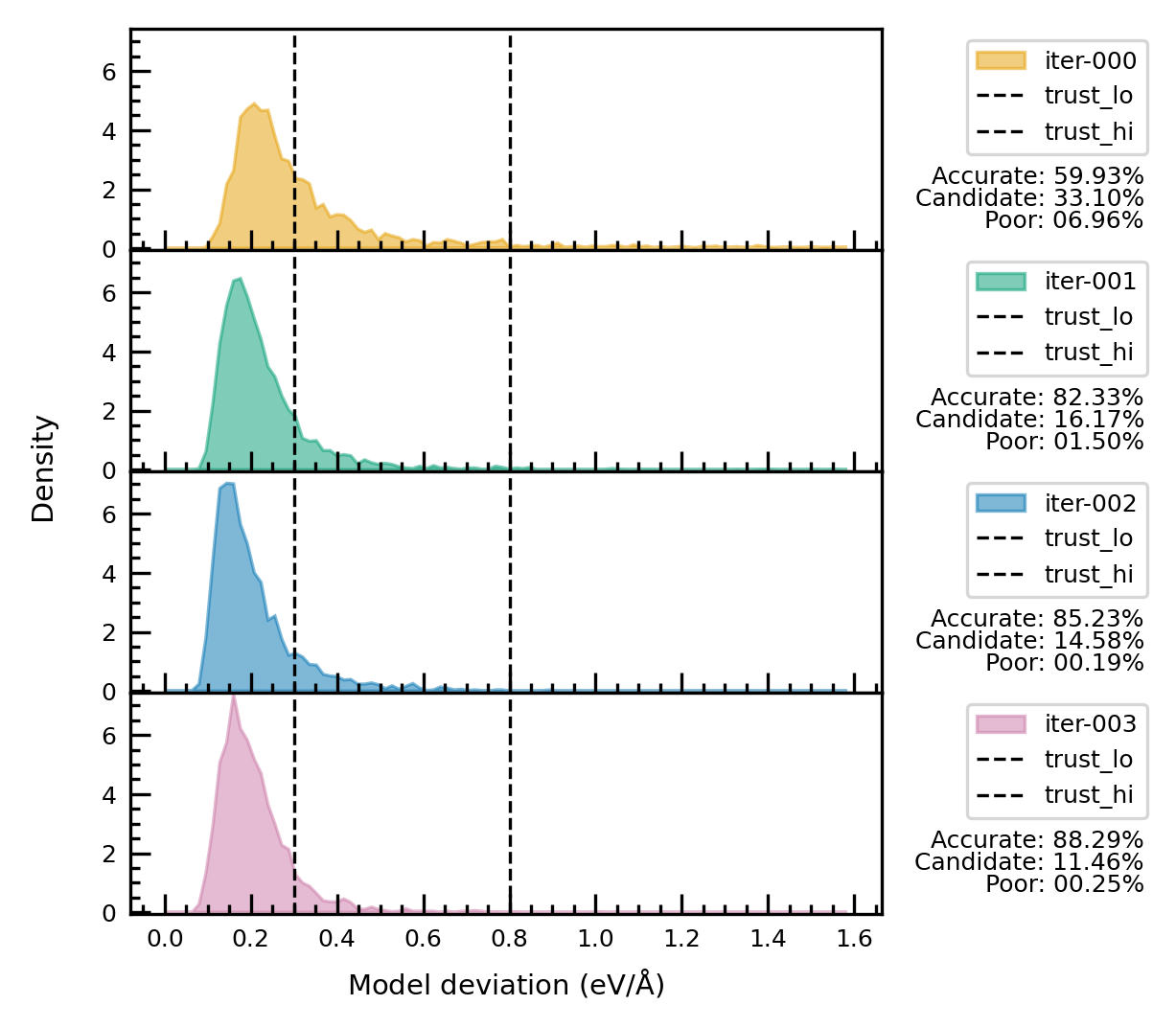
Plot model_devi.out#
function for plotting many model_devi.out files together.
from ectoolkits.plots.dpmd import plot_model_devi
trust_lo = 0.30
trust_hi = 0.80
prefix = Path("path_to_model_devi_files/")
# general structures of model_devi_files
# [
# [ model_devi_file1, file2 ,... ], # iteration 0
# [ model_devi_file1, file2 ,... ], # iteartion 1
# ]
# all files in one iteration are merged
model_devi_files = [
[
prefix/f"iter-{i_iter:03d}/model_devi-{i_model_devi}.out" for i_model_devi in range(4)
]
for i_iter in range(4)
]
# Call the function
fig = plot_model_devi(model_devi_files, trust_lo, trust_hi, bins=100, density=True, range=(0, 1.6))